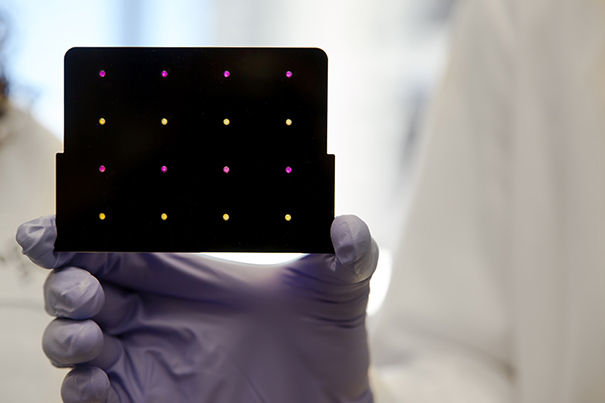
A black cartridge containing a paper-based diagnostic for detecting the Zika virus is held up by a researcher at Harvard’s Wyss Institute. Areas that turn purple indicate samples infected with Zika, while yellow areas indicate samples that are free of the virus.
Photo courtesy of Wyss Institute at Harvard University
Paper disc can quickly detect Zika virus in the field
Inexpensive method could slow spread of outbreak, future pandemic diseases
Researchers have developed a system that can quickly diagnose a patient with Zika, according to synthetic biologist James Collins of the Wyss Institute for Biologically Inspired Engineering at Harvard University.
The low-cost, rapid paper-based diagnostic tool screens blood, urine, or saliva samples for specific strains of the virus, said Collins, who led the international, multi-institutional team of researchers.
“The growing global health crisis caused by the Zika virus propelled us to leverage novel technologies we have developed in the lab and use them to create a workflow that could diagnose a patient with Zika, in the field, within two to three hours,” he said.
Collins is a Wyss core faculty member and Termeer Professor of Medical Engineering & Science and professor of biological engineering at Massachusetts Institute of Technology’s Department of Biological Engineering.
Building on previous work done at Harvard’s Wyss Institute by Collins and his team, joined with collaborators from MIT, the Broad Institute of Harvard and MIT, Harvard Medical School, University of Toronto, Arizona State University (ASU), University of Wisconsin-Madison, Boston University, Cornell University, and Addgene to prototype the rapid diagnostic test and describe their methods in a study published online today in the journal Cell — all within six weeks. Collins is the paper’s corresponding author.
Emerging innovation during the Ebola health crisis
In October 2014, Collins’ team developed a breakthrough method for embedding synthetic gene networks — which could be used as programmable diagnostics and sensors — on portable, small discs of ordinary paper.
Stirred by the then-ongoing Ebola outbreak in Africa, they demonstrated a proof-of-concept color-changing diagnostic that could screen for the disease by embedding in paper a synthetic biomolecular sensor designed to detect specific RNA sequences. These RNA sequences can mark not only the genetic signatures of Ebola but also other RNA viruses, including Zika, SARS, measles, influenza, hepatitis C, and West Nile fever.
The team believed that the method could be applied in the field to identify viruses with blood, urine, or saliva samples. However, until recently, the paper-based technology was challenged by the extremely low concentrations of virus normally found in these body fluids. Now, using blood samples from monkeys infected with Zika as well as virus recovered from cells infected in the laboratory, the team has validated a next-generation technique that overcomes this problem.
More like this
A leap for paper-based diagnostics, urged by the Zika outbreak
“The vivid images in the news stemming from the ongoing Zika crisis are heartbreaking,” said Keith Pardee, one of the study’s co-first authors and an assistant professor in the Leslie Dan Faculty of Pharmacy at University of Toronto. “We hope a tool like this can help reduce the impact of the outbreak until a vaccine can be developed.”
Collins’ team designed a three-step workflow: amplification, Zika detection, and CRISPR-Cas9-aided strain identification. CRISPR-Cas9, a gene-editing mechanism derived from the immune systems of bacteria, can be used to search entire sequences to find exclusive genetic markers. Leveraging CRISPR-Cas9’s talent for sequence recognition, the third part of the team’s system uses a CRISPR-Cas9-aided paper-based diagnostic to discriminate between strains whose genetic profiles differ by as little as one nucleotide.
Once a sample’s RNA has been amplified using a mixture of enzymes and “primers” (DNA sequences that trigger replication), a drop is administered to paper discs that are freeze-dried containing a mixture of cellular components and biological proteins. The droplet of amplified RNA activates the freeze-dried components so that the discs change color to indicate a positive result for Zika virus. While the result can be read with the naked eye, similar to a home pregnancy test, a specially designed electronic reader can get faster results and could, one day, quantify the amount of viral load in a sample.
If Zika is detected, the third step involves mixing a sample with a freeze-dried CRISPR-Cas9 cocktail and using that mixture to wet another set of color-changing paper discs. Depending on the strain of Zika contained in the sample, these discs undergo another set of visible color changes.
“We have tested our diagnostic systems against closely related strains of the Dengue virus and found that within the first two steps, our system can readily distinguish Zika from Dengue,” said Alexander Green, co-first author on the study and an assistant professor in the Center for Molecular Design and Biomimetics at ASU’s Biodesign Institute and School of Molecular Sciences.
Combating future pandemics
All components of the diagnostic system can be freeze-dried for storage and transport while retaining their efficacy.
“In response to an emerging outbreak, we envision a custom-tailored diagnostic system could be ready for use within one week’s time,” said Collins.
“The ability to recapitulate the genetic machinery of living cells in ordinary freeze-dried paper provides a way to develop revolutionary sensors and diagnostics in a fraction of the time and with higher sensitivity and specificity than conventional assays. These inexpensive paper-based tests also can be easily transported out of the laboratory and distributed virtually anywhere in the world.
“The potential for applications in health and environmental screening, particularly in low-resource areas, is huge,” said Wyss Institute director Donald Ingber, who is the Judah Folkman Professor of Vascular Biology at Harvard Medical School and the Vascular Biology Program at Boston Children’s Hospital, and also professor of bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
The work was supported by the Wyss Institute for Biologically Inspired Engineering, MIT’s Center for Microbiome Informatics and Therapeutics, the Defense Threat Reduction Agency, Arizona State University (A.G.), and the National Institutes of Health.
To read the full release, visit the Wyss Institute website.





