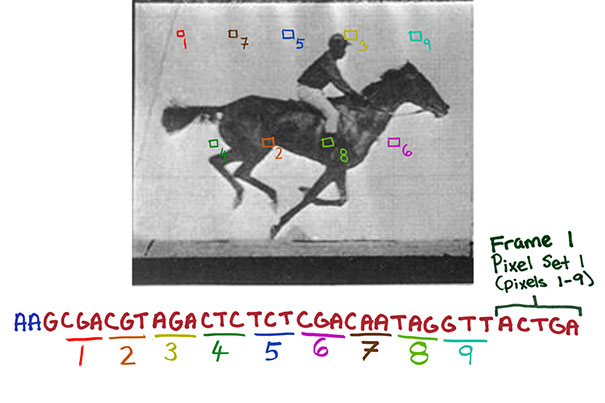
CRISPR system-based technology lets researchers record digital data — such as successive frames of a movie of a galloping horse — in a population of living bacteria. This could eventually have cells record the key changes they undergo during development or exposure to environmental or pathogenic signals.
Credit: Wyss Institute at Harvard University
New CRISPR technology takes cells to the movies
Genome engineering system transforms living cells into archival data storage devices
Researchers use expensive machinery to develop ways to harness DNA as a synthetic raw material to store large amounts of digital information outside of living cells.
But what if they could coerce living cells, such as large populations of bacteria, to use their own genomes as a biological hard drive that can record information scientists could tap anytime? That approach not only could open entirely new possibilities of data storage, it could also be engineered into an effective memory device able to create a chronological record of cells’ molecular experiences during development or under exposure to stresses or pathogens.
In 2016, a team at the Wyss Institute for Biologically Inspired Engineering and Harvard Medical School (HMS) led by Wyss core faculty member George Church built the first molecular recorder based on the CRISPR system. The recorder allows cells to acquire bits of chronologically provided, DNA-encoded information that generate a memory in a bacterium’s genome. The information is stored as an array of sequences in the CRISPR locus and can be recalled and used to reconstruct a timeline of events.
“As promising as this was, we did not know what would happen when we tried to track about 100 sequences at once, or if it would work at all. This was critical since we are aiming to use this system to record complex biological events as our ultimate goal,” said Seth Shipman, a postdoctoral fellow working with Church and the study’s first author.
Now they know. In a study published today in Nature, the same team shows in foundational proof-of-principle experiments that developed further as a first-of-its-kind approach, the CRISPR system can encode information in living cells that is as complex as a digitized image of a human hand, reminiscent of early humans’ paintings on cave walls and a sequence of one of the first motion pictures made ever, Eadweard Muybridge’s film of a galloping horse.
The CRISPR system helps bacteria develop immunity against the constant onslaught of viruses in their environments. As a memory of survived infections, it captures viral DNA molecules and generates short “spacer” sequences from them, which it then adds as new elements upstream of previous elements in a growing array located in the bacterial genomes’ CRISPR locus. The CRISPR-Cas9 protein uses this memory to destroy the same viruses when they return. But other than Cas9, now famous as a widely used genome-engineering tool, other parts of the CRISPR system so far have not been much exploited.
“In this study, we show that two proteins of the CRISPR system, Cas1 and Cas2, that we have engineered into a molecular recording tool, together with new understanding of the sequence requirements for optimal spacers, enables a significantly scaled-up potential for acquiring memories and depositing them in the genome as information that can be provided by researchers from the outside, or that, in the future, could be formed from the cells natural experiences,“ said Church, the Robert Winthrop Professor of Genetics at Harvard Medical School and a Professor of Health Sciences and Technology at Harvard and MIT.
“Harnessed further, this approach could present a way to cue different types of living cells in their natural tissue environments into recording the formative changes they are undergoing into a synthetically created memory hotspot in their genomes,” he said.
The team used still and moving images because they represent constrained and clearly defined data sets; the movie also gave the bacteria a chance to acquire information frame by frame.
“We designed strategies that essentially translate the digital information contained in each pixel of an image or frame as well as the frame number into a DNA code, that, with additional sequences, is incorporated into spacers. Each frame thus becomes a collection of spacers,” Shipman said. “We then provided spacer collections for consecutive frames chronologically to a population of bacteria which, using Cas1/Cas2 activity, added them to the CRISPR arrays in their genomes. And after retrieving all arrays again from the bacterial population by DNA sequencing, we finally were able to reconstruct all frames of the galloping horse movie and the order they appeared in.”
Shipman and postdoctoral fellow Jeff Nivala, the study’s second author, defined a set of requirements they expect will make the spacer sequences easier to acquire, as well as sequence features that prevent their acquisition into growing CRISPR arrays.
In future work, the team will focus on establishing molecular recording devices in other cell types and on engineering the system to memorize biological information.
“One day, we may be able to follow all the developmental decisions that a differentiating neuron is taking from an early stem cell to a highly-specialized type of cell in the brain, leading to a better understanding of how basic biological and developmental processes are choreographed,” said Shipman. Ultimately, the approach could lead to better methods for generating cells for regenerative therapy, disease modeling, and drug testing.
The study was supported by grants from the National Institute of Mental Health, the National Human Genome Research Institute, the Simons Foundation Autism Research Initiative, the National Institute of Neurological Disorders and Stroke, Paul G. Allen Frontiers Group, and the Wyss Institute.