A zebrafish embryo develops.
Video by Fengzhu Xiong and Sean Megason/HMS
From one, many
Harvard scientists reveal the genetic roadmap to building an entire organism from a single cell
Whether a worm, a human, or a blue whale, all multicellular life begins as a single-celled egg.
From this solitary cell emerges the galaxy of others needed to build an organism, with each new cell developing in the right place at the right time to carry out a precise function in coordination with its neighbors.
This feat is one of the most remarkable in the natural world, and despite decades of study, a complete understanding of the process has eluded biologists.
Now, in three landmark studies published online today in Science, Harvard Medical School and Harvard University researchers report how they have systematically profiled every cell in developing zebrafish and frog embryos to establish a roadmap revealing how one cell builds an entire organism.
Using single-cell sequencing technology, the research teams traced the fates of individual cells over the first 24 hours of the life of an embryo. Their analyses reveal the comprehensive landscape of which genes are switched on or off, and when, as embryonic cells transition into new cell states and types.
Together, the findings represent a catalog of genetic “recipes” for generating different cell types in two important model species and provide an unprecedented resource for the study of developmental biology and disease.
“With single-cell sequencing, we can, in a day’s work, recapitulate decades of painstaking research on the decisions cells make at the earliest stages of life,” said Allon Klein, HMS assistant professor of systems biology and co-corresponding author of two of the three Science studies.
Biomedically, these baseline resources for how organisms develop are as important as having baseline resources for their genomes, the researchers said.
“With the approaches that we’ve developed, we’re charting what we think the future of developmental biology will be as it transforms into a quantitative, big-data-driven science,” Klein said.
In addition to shedding new light on the early stages of life, the work could open the door to a new understanding of a host of diseases, said Alexander Schier, the Leo Erikson Life Sciences Professor of Molecular and Cellular Biology at Harvard, and a corresponding author of the third study.
“We foresee that any complex biological process in which cells change gene expression over time can be reconstructed using this approach,” Schier said. “Not just the development of embryos but also the development of cancer or brain degeneration.”
A Xenopus egg divides.
Courtesy of Enrique Amaya
One at a time
Every cell in a developing embryo carries within it a copy of the organism’s complete genome. Like construction workers using only the relevant portion of a blueprint when laying a building’s foundation, cells must express the necessary genes at the appropriate time for the embryo to develop correctly.
In their studies, Klein collaborated with co-authors Marc Kirschner, the HMS John Franklin Enders University Professor of Systems Biology, Sean Megason, HMS associate professor of systems biology, and colleagues to analyze this process in zebrafish and western claw-toed frog (Xenopus tropicalis) embryos, two of the most well-studied model species in biology.
The researchers leveraged the power of InDrops, a single-cell sequencing technology developed at HMS by Klein, Kirschner, and colleagues to capture gene-expression data from each cell of the embryo, one at a time. The teams collectively profiled more than 200,000 cells at multiple time points over 24 hours from both species.
To map the lineage of essentially every cell as an embryo develops, along with the precise sequence of gene-expression events that mark new cell states and types, the teams developed new experimental and computational techniques, including the introduction of artificial DNA bar codes to track the lineage relationships between cells, called TracerSeq.
“Understanding how an organism is made requires knowing which genes are turned on or off as cells make fate decisions, not just the static sequence of a genome,” Megason said. “This is the first technological approach that has allowed us to systematically and quantitatively address this question.”
“It is almost like going from seeing a few stars to seeing the entire universe.”
Alexander Schier, the Leo Erikson Life Sciences Professor of Molecular and Cellular Biology
In the study co-led by Schier, the research team used Drop-Seq — a single-cell sequencing technology developed by researchers at HMS and the Broad Institute of MIT and Harvard — to study zebrafish embryos over 12 hours at high time resolution. Teaming with Aviv Regev, a core member at the Broad, the team reconstructed cell trajectories through a computational method they named URD, after the Norse mythological figure who decides all fates.
Schier and colleagues profiled more than 38,000 cells, and developed a cellular “family tree” that revealed how gene expression in 25 cell types changed as they specialized. By combining that data with spatial inference, the team was also able to reconstruct the spatial origins of the various cells types in the early zebrafish embryo.
Recipe for success
In both species, the teams’ findings mirrored much of what was previously known about the progression of embryonic development, a result that underscored the power of the new approaches. But the analyses were unprecedented in revealing in comprehensive detail the cascades of events that take cells from early progenitor or “generalist” states to more specialized states with narrowly defined functions.
The teams identified otherwise difficult-to-detect details such as rare cell types and subtypes and linked new and highly specific gene-expression patterns to different cell lineages. In several cases, they found cell types emerging far earlier than previously had been thought.
For scientists striving to answer questions about human disease, these data could be powerfully illuminating. In regenerative medicine, for example, researchers have for decades aimed to manipulate stem cells toward specific fates with the goal of replacing defective cells, tissues, or organs with functional ones. Newly gleaned details about the sequence of gene expression changes that precipitate the emergence of specific cell types can propel these efforts further.
“With these datasets, if someone wants to make a specific cell type, they now have the recipe for the steps that those cells took as they formed in the embryo,” Klein said. “We’ve in some sense established a gold standard reference for how complex differentiation processes actually progress in embryos, and set an example for how to systematically reconstruct these types of processes.”
When combined with one of the core concepts in biological inquiry — the idea of disrupting a system to study what happens — single-cell sequencing can yield insights difficult to attain before, Klein said.
As a proof of principle, Klein, Megason, and colleagues used the CRISPR/Cas9 gene editing system to create zebrafish with a mutant form of chordin, a gene involved in determining the back-to-front orientation of a developing embryo. Schier and colleagues took a similar approach by profiling zebrafish with a mutation in a different patterning gene known as one-eyed pinhead.
When analyzed with single-cell sequencing, the teams confirmed previously known descriptions of chordin and one-eyed pinhead mutants, and could describe in detail or even predict the effects of these mutations on developing cells and nascent tissues across the whole embryo.
Unexpectedly, the groups independently found that at the single-cell level, gene expression was the same in mutants and wild-type, despite the loss of an essential signaling pathway. The proportions of different cell types, however, changed.
“This work only became possible through recent technologies that let us analyze gene expression in thousands of individual cells,” Schier said. “Now the scale is much larger, so that we can reconstruct the trajectory of almost all cells and all genes during embryogenesis. It is almost like going from seeing a few stars to seeing the entire universe.”
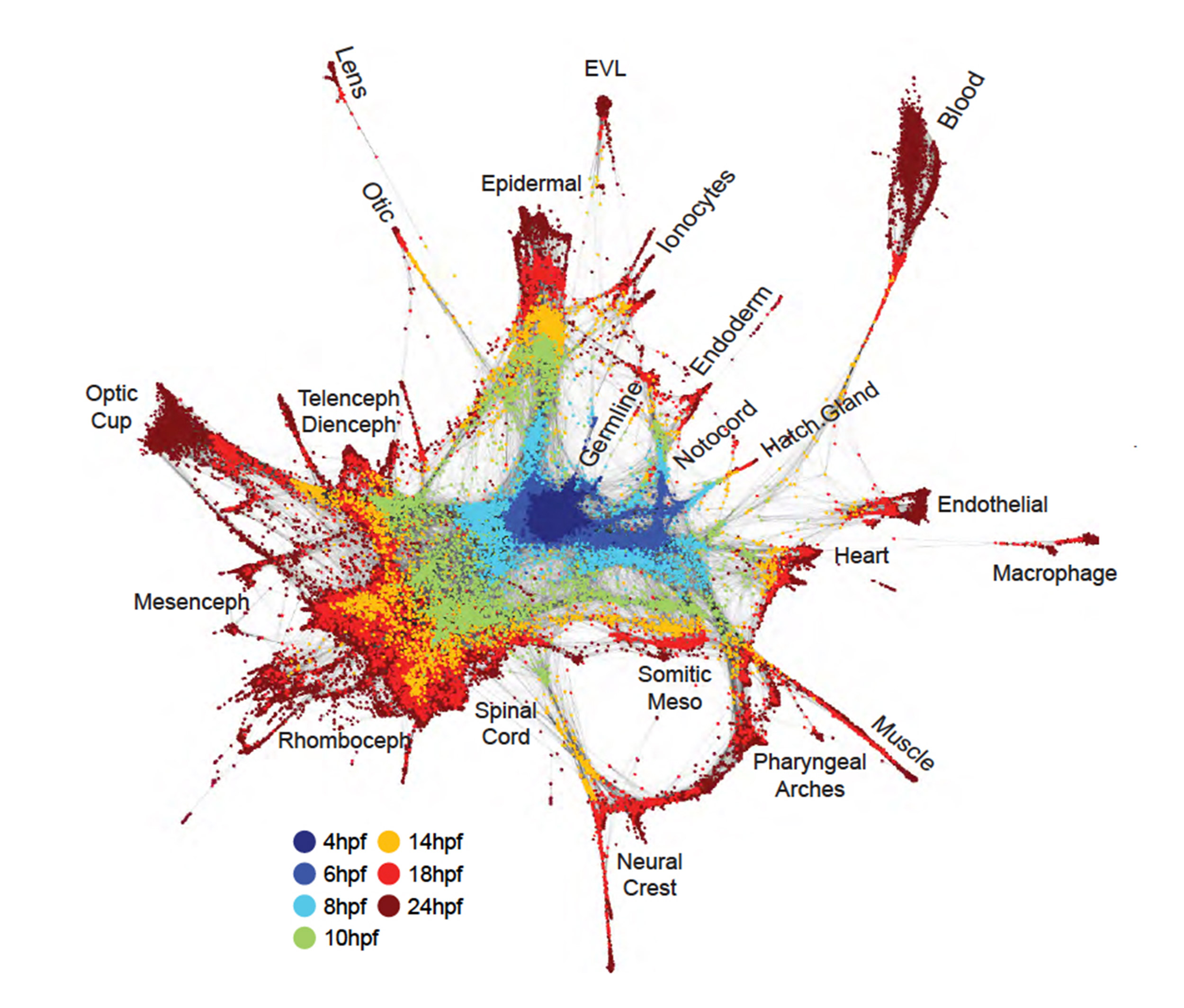
Rethinking definitions
The research teams also demonstrated how these data can be mined to answer long-standing fundamental questions in biology.
When Klein, Kirschner, Megason, and colleagues compared cell-state landscapes between zebrafish and frog embryos, they observed mostly similarities. But their analyses revealed numerous surprises as well. One such observation was that genes marking cell states in one species were often poor markers for the same cell state in the other species.
In several instances, they found that the DNA sequence of a gene — and the structure of the protein it encoded — could be nearly identical between species but have very different expression patterns.
“This really shocked us, because it goes against all the intuition we had about development and biology,” Klein said. “It was a really uncomfortable observation. It directly challenges our idea of what it means to be a certain ‘cell type.’”
The reason these differences were not spotted before, the researchers hypothesize, is that computational analyses “pay attention” to data in a way fundamentally different from how humans do.
“I think this reflects some level of confirmation bias. When scientists find something conserved between species, they celebrate it as a marker,” Megason said. “But often, all the other nonconserved features are ignored. Quantitative data helps us move past some of these biases.”
In another striking finding, the teams observed that the process of cell differentiation into distinct types — which is commonly thought to occur in a tree-like structure where different cell types branch off from a common ancestor cell — can form “loops” as well as branches.
For example, the neural crest — a group of cells that give rise to diverse tissue types including smooth muscle, certain neurons, and craniofacial bone — initially emerges from neural and skin precursors, but is well-known to generate cells that appear almost identical to bone and cartilage precursors.
The new results suggest that similar loops might occur in other situations. That cells in the same state can have very different developmental histories suggests that our hierarchical view of development as a “tree” is far too simplified, Klein said.
All three teams also identified certain cell populations that existed in a kind of intermediate “decision-making” state. Schier and colleagues found that, at certain key developmental branch points, cells appeared to go down one developmental trajectory but then changed their fate to another.
Klein, Megason, Kirschner, and colleagues made a related observation that, early in development, some cells activated two distinct developmental programs. Though those intermediate cells would eventually adopt a single identity, these discoveries add to the picture of how cells develop their fate and hint that there may be factors beyond genes involved in directing that.
“With multilineage cells, we have to start wondering if their final fate is being determined by some selective force or interaction with the environment, rather than just genetic programs,” Kirschner said.
Future foundation
The newly generated data sets and the new tools and technologies developed as part of these studies lay the foundation for a wide spectrum of future exploration, according to the authors.
Developmental biologists can gather more and higher-quality data on many species, follow embryos further in time, and perform any number of perturbation experiments, all of which can help improve understanding of the fundamental rules of biology and disease.
These resources can also serve as a focal point for collaboration and interaction since most labs do not have the depth of expertise needed to exploit all the data and information generated, the authors noted.
“I think these studies are creating a real sense of community, with researchers raising questions and interacting with each other in a way that harkens back to earlier times in the study of embryology,” Kirschner said.
The three studies, Schier said, are an example of how the scientific community can work on complementary questions to answer important questions in biology.
“Instead of competing, our groups were in regular contact over the past two years and coordinated the publication of our studies,” he said. “And it is great how complementary the three papers are — each highlights different ways such complex data sets can be generated, analyzed and interpreted.”
The next conceptual leap, the teams suggest, will be to better understand how cell-fate decisions are made.
“Right now, we have a roadmap, but it doesn’t tell us what the signs are,” Megason said. “What we need to do is figure out the signals that direct cells down certain roads, and what the internal mechanisms are that allow cells to make those decisions.”
Whatever the future holds, these data sets will leave their mark.
“The beauty of working on an organism is that this is it,” Klein said. “Ten, 20 years from now, we can still be sure zebrafish and frogs are going to develop according to the same patterns.”
All three research teams have made their data sets and tools available as interactive, browsable online resources. For Klein and colleagues: zebrafish and Xenopus. For Schier and colleagues: zebrafish and URD.
Daniel Wagner is the lead author and Sean Megason and Allon Klein are co-corresponding authors of the study “Systematic mapping of cell state trajectories, cell lineage, and perturbations in the zebrafish embryo using single cell transcriptomics.” Additional authors are Caleb Weinreb, Zach Collins, and James Briggs. The study was supported by the National Institutes of Health (1K99GM121852, R01GM107733, R01DC015478), the Edward J. Mallinckrodt, Jr. Foundation and the Burroughs Wellcome Fund.
James Briggs is the lead author and Marc Kirschner and Allon Klein are co-corresponding authors of the study “The dynamics of gene expression in vertebrate embryogenesis at single cell resolution.” Additional authors are Caleb Weinreb, Daniel Wagner, Sean Megason, and Leonid Peshkin. The study was supported by the National Institutes of Health (R21HD087723), the Edward J. Mallinckrodt Foundation and the Burroughs Wellcome Fund.
Jeffrey Farrell and Yiqun Wang are co-lead authors and Aviv Regev and Alexander Schier are co-corresponding authors of the study “Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis.” Additional authors are Samantha Riesenfeld and Karthik Shekhar. The study was supported by the National Institutes of Health, the Allen Discovery Center for Cell Lineage Tracing, the Jane Coffin Childs Memorial Fund, the Charles A. King Trust, the Howard Hughes Medical Institute, and the Klarman Cell Observatory.





