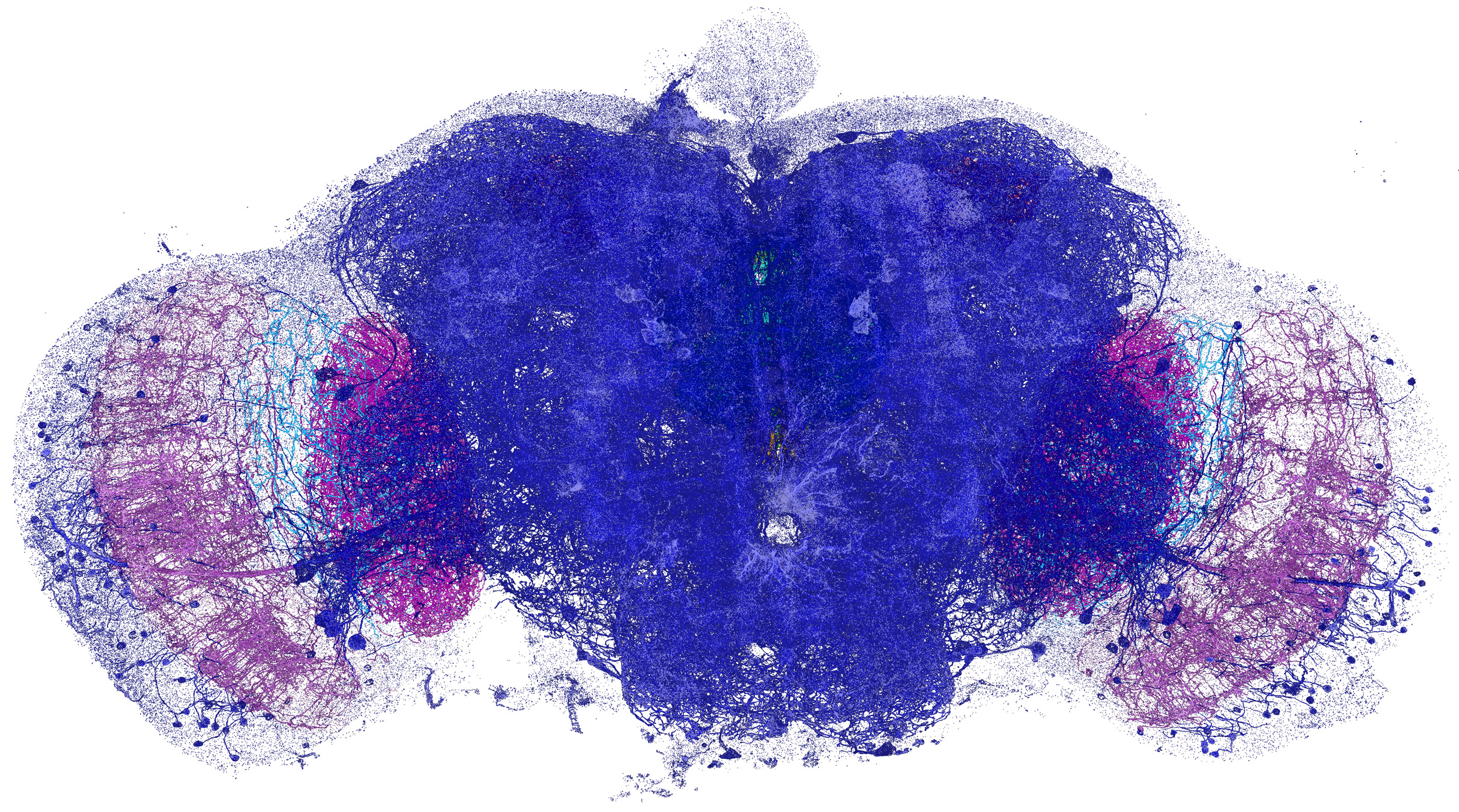
Neural processes (yellow) and pre- and post-synaptic proteins (teal and magenta) in a mouse brain.
Video and images courtesy of Gao et al/Science 2019
Science at the speed of ‘light-sheet’
New technique enables subcellular imaging of brain tissue 1,000 times faster than other methods
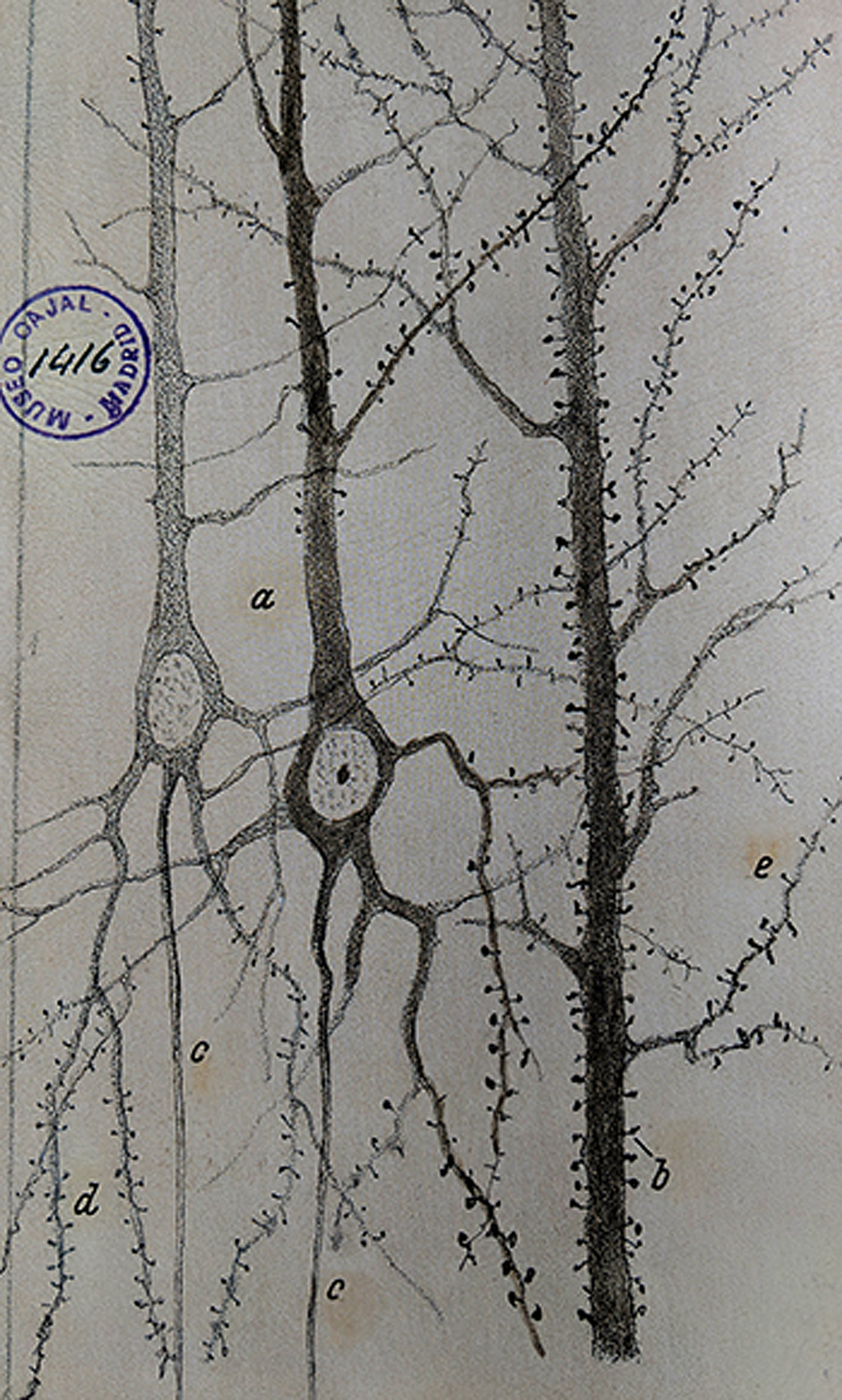
In the late 19th century, the Spanish anatomist Santiago Ramón y Cajal laid the foundation for modern neuroscience with a microscope, a pen, and some paper. Applying a cell-staining technique to samples of brain tissue, he produced thousands of detailed illustrations that revealed for the first time the intricate complexity of neurons and neuronal networks. Based on his observations, Ramón y Cajal proposed that the neuron was the basic functional unit of the nervous system, a hypothesis confirmed when the electron microscope was invented in the 1950s.
In the decades since, microscopy has remained central to efforts to understand the brain in health and disease. Scientists today can visualize brain tissues in remarkable detail, down to the level of the individual proteins responsible for neuronal structure and function. Despite technological advances, however, challenges remain. Attempts to achieve that level of resolution for large samples can lead to tissue damage or be prohibitively time-consuming — imaging a single whole fruit fly brain with an electron microscope, for example, can require years or even decades of work.
Now, researchers at Howard Hughes Medical Institute’s Janelia Research Campus, the Massachusetts Institute of Technology, and Harvard Medical School have developed a method that yields high-resolution visualizations of large volumes of brain tissue at speeds roughly 1,000 times faster than other methods. Combining two recently developed technologies — expansion microscopy and lattice light-sheet microscopy — enabled them to image an entire fruit fly brain, as well as large sections of mouse cortex, in subcellular detail in only a few days.
The work — led by HHMI investigators Eric Betzig of Janelia and Edward Boyden of MIT, and carried out in collaboration with Srigokul Upadhyayula, HMS assistant professor of pediatrics at Boston Children’s Hospital, and Tomas Kirchhausen, HMS professor of cell biology in the Blavatnik Institute at HMS — was detailed in a study published online in Science on Jan. 17.
“The greatest scientific challenge of the 21st century is the brain, perhaps the most complex structure that we know of in the universe,” said Upadhyayula, who is co-first author of the study. “This new method can potentially usher in a new era for neuroscience, as we explore the ultrafine architecture of neurons and neural circuits across the brain, at scales and resolutions that were previously unattainable.”
Rapidly imaging large volumes of brain tissue at high resolution could lead to detailed maps of the activity and wiring of the brain, the authors said, which may help scientists better understand diseases of the brain, build better artificial intelligence, or even explain the molecular drivers of behavior, decision-making, and more.
“If we think of the brain as a city, we’ve only been able to look at small neighborhoods in detail. But now, we have the ability to create something like Google Maps,” said Kirchhausen, the HMS Springer Family Professor of Pediatrics at Boston Children’s.
“This approach could let us walk around virtually, zoom into any building or neighborhood we’re interested in, zoom out to see how they are connected, and much more,” Kirchhausen said. “We can explore the city in its entirety at an exquisite level of detail, making discoveries and hopefully explaining the previously unknown.”
Crystal clear
Work on the project began when co-first authors Ruixuan Gao and Shoh Asano of the Boyden lab decided to image complete neural circuits spanning millimeters or more using expansion microscopy. This technique, developed by Boyden and colleagues, enlarges small specimens so that molecular details can be seen more easily with conventional optical microscopes.
To do it, tissue samples are infused with swellable gels, similar to what’s found in diapers. The molecules within the tissue lock into the gel scaffold, keeping the same relative positions in 3-D even after expansion. Because 3-D expansion dramatically increases the volume of a sample, the team needed a microscope that was high-resolution, high-speed, and gentle enough that it wouldn’t photo-bleach or otherwise damage the sample before it could be imaged in its entirety.
The solution was lattice light-sheet microscopy, developed by Betzig and colleagues, which can rapidly image tissues at subcellular resolutions. By sweeping an ultrathin sheet of light through a sample and illuminating only what lies in the microscope’s plane of focus, the sample remains undamaged.
“The result is that we get crystal-clear images at blazingly fast speeds over very large volumes compared to earlier microscopy techniques,” Boyden said.
The team’s first tests combining the techniques in expanded mouse brain tissues were almost immediately successful, revealing intricate subcellular details such as dendritic spines that protrude from neurons like leaf buds on a tree branch in spring.
“I couldn’t believe the quality of the data I was seeing. You could have knocked me over with a feather,” said Betzig, now a professor at the University of California, Berkeley.
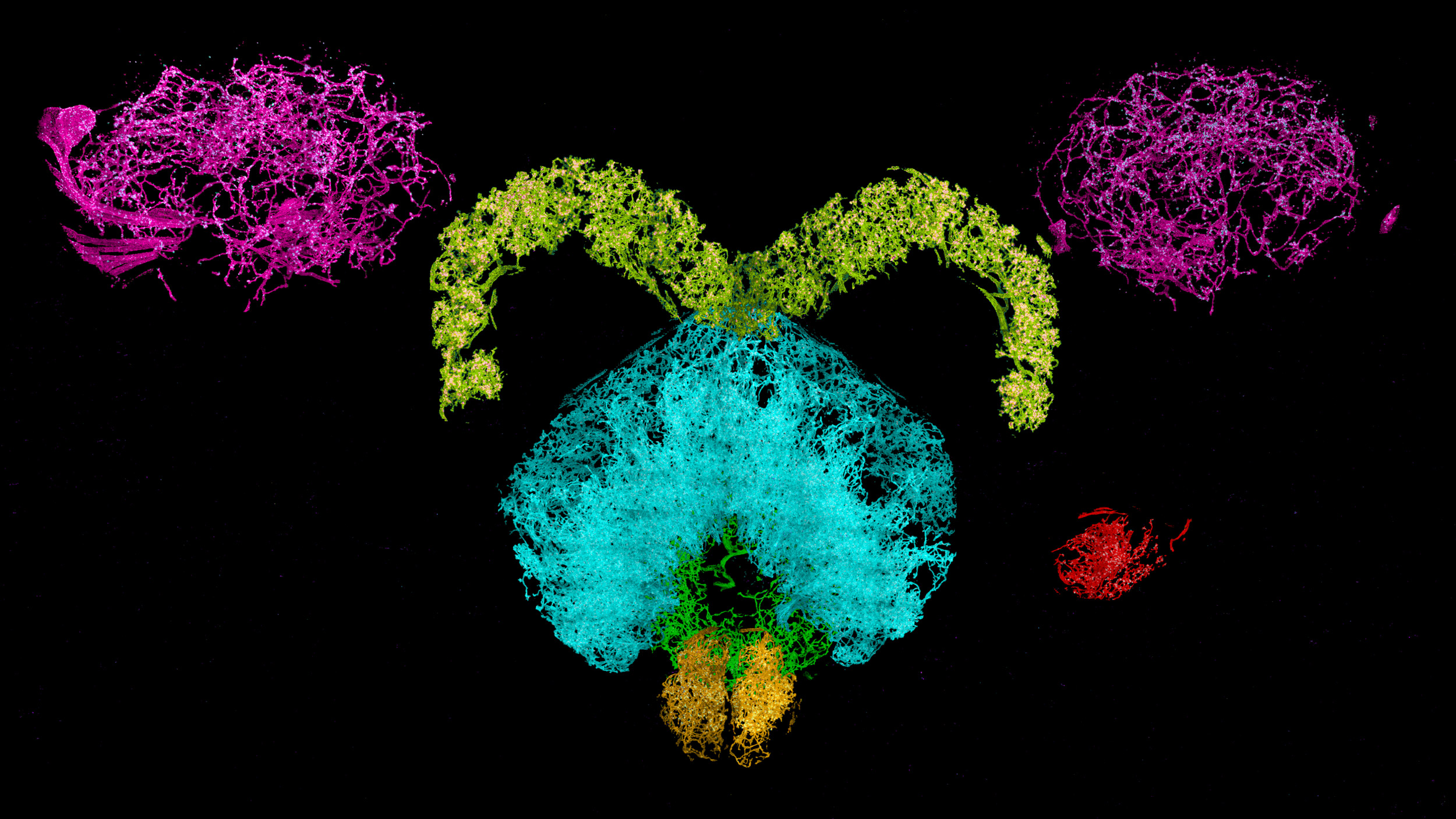
Dopaminergic neurons and associated synaptic proteins in a fruit fly brain.
Avengers assemble
Over the past two years, the team of biologists, microscopists, physicists, and computer scientists has worked together to capture and analyze images of sections of mouse cortex and entire fruit fly brains.
“This is like Avengers-level collaboration,” said Gao, referring to the comic book superhero team.
The new approach is data-intensive. To visualize the 20-million-cubic-micrometer volume of a whole fruit fly brain, for example, the team first had to image more than 51,000 3-D subsections of the brain, representing roughly 10 terabytes of data. These data were then computationally stitched back together like a 3-D jigsaw puzzle.
Upadhyayula led the development of bespoke algorithms to analyze the data and generate stunning movies that show the brain’s intricacies. Together, the team studied more than 1,500 dendritic spines, imaged fatty sheaths that insulate mouse nerve cells, highlighted all the dopaminergic neurons, and counted all the roughly 40 million synapses across the entire fly brain.
The technology also gives the researchers the ability to tag and trace specific proteins through the brain, allowing them to ask new questions about structure and function.
“In our study, we barely scratched the surface in terms of the depth of information that’s buried in this incredible wealth of data,” Upadhyayula said. “We’re now working to reveal and present all of that information in a way that will let us ask questions that we couldn’t ask before because of the lack of throughput at this level of resolution.”
One of the technique’s most exciting aspects, the authors said, is that it lets them explore in molecular detail brain-to-brain differences in neurons and neuronal networks — young versus old, male versus female, healthy versus diseased. It can also enable visualizations and new lines of research in other organs, biological systems, and a variety of diseases including cancer, the scientists said.
“Ramón y Cajal showed us the beauty and complexity of neurons, and we are essentially expanding his work to the complete volume of a brain,” Kirchhausen said. “This is important because we’re still at the stage where every time we learn something new about the brain or any other biological system, it holds consequence — whether for our understanding of ourselves, or for translational and clinical applications.
“To make serious advances, we need people who are fluent in computation and physics as much as we need biologists, and it is essential that we work as a team. I think this project is an incredible demonstration of what can be achieved through exactly such teamwork.”
Additional authors include Igor Pisarev, Daniel E. Milkie, Tsung-Li Liu, Ved Singh, Austin Graves, Grace H. Huynh, Yongxin Zhao, John Bogovic, Jennifer Colonell, Carolyn M. Ott, Christopher Zugates, Susan Tappan, Alfredo Rodriguez, Kishore R. Mosaliganti, Shu-Hsien Sheu, H. Amalia Pasolli, Song Pang, C. Shan Xu, Sean G. Megason, Harald Hess, Jennifer Lippincott-Schwartz, Adam Hantman, Gerald M. Rubin, Stephan Saalfeld, and Yoshinori Aso.
The study, “Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution,” was supported by HHMI, the National Institutes of Health (1R01NS087950, 1RM1HG008525, 1R01DA045549, 2R01DA029639, 1R01NS102727, 1R41MH112318, 1R01EB024261, 1R01MH110932, R01DC015478, R44MH093011, 1DP1NS087724, R01GM075252), John Doerr, the Open Philanthropy Project, IARPA D16PC00008, the U.S. Army Research Laboratory and the U. S. Army Research Office, U.S.-Israel Binational Science Foundation, Biogen, and Ionis Pharmaceuticals.
Adapted from an HHMI news release.





